cellularcommunication
our research IS driven by both fundamental and clinical questions intersecting the fields of tumour biology, immunology and regeneration. we work to identify pathways of (INTRA) cellular communication that can be exploited to selectively target cells in both cancer and (auto)immunity.
CURRENT research areas include:
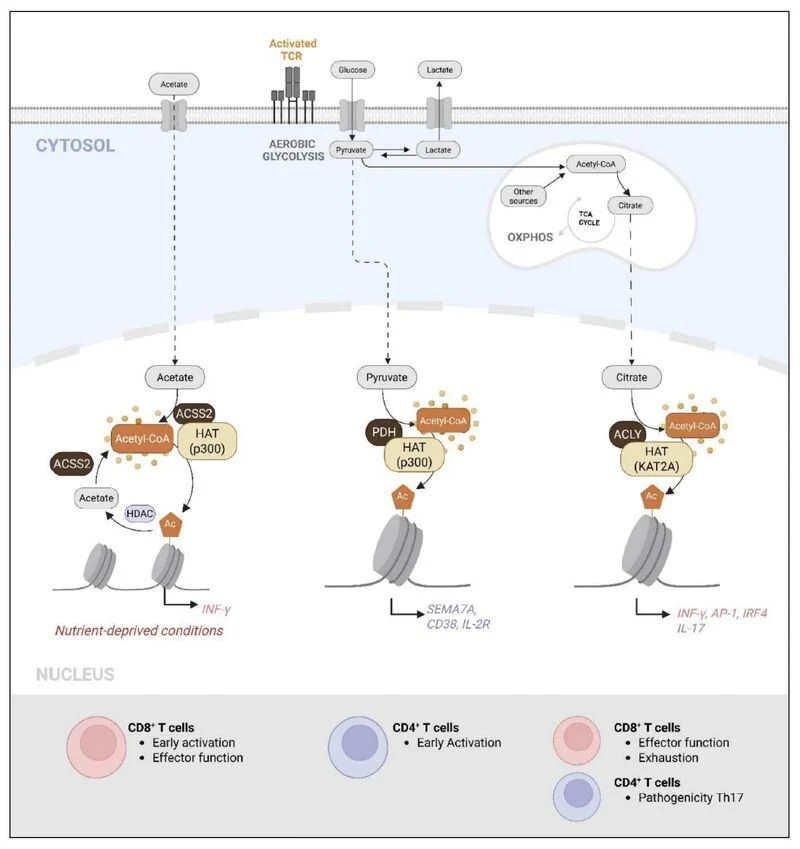
- UNDERSTANDING HOW INTRACELLULAR METABOLISM CONTROLS CHROMATIN REMODELLING AND TRANSCRIPTIONAL PROGRAMMING
-understanding how developmental signals and intracellular metabolism interact to drive somitogenesis
- characterizing molecular pathways controlling t cell activation, anergy and autophagy.
- defining and manipulating INTERACTIONS BETWEEN IMMUNE CELLS & TUMOURS
BIOLOGICAL PROCESSES WE explore INCLUDE: TRANSCRIPTION, intracellular SIGNALING, EPIGENETIC regulation, cellular METABOLISM, and AUTOPHAGY
Our research uses a broad range of state-of-the-art molecular and cell biological approaches, as well as in vivo and ex-vivo models of disease. This forms a translatable research program with close connection to our clinical collaborators.
We located at the center for molecular medicine and part of regenerative medicine utrecht.
Phone
+31 (0)30 212-1800
Location
Regenerative Medicine Center
Hubrecht Location
UMC Utrecht
Uppsalalaan 8
3584CT Utrecht
Publications highlighting current research themes:
Glycolytic reprogramming shapes the histone acetylation profile of activated CD4+ T cells in juvenile idiopathic arthritis. Mocholi E, Corrigan E, Chalkiadakis T, Gulersommez C, Stigter E, Vastert B, van Loosdregt J, Prekovic S, Coffer PJ (2025).
These findings provide support for targeting metabolism to reshape the epigenome of inflammatory T cells and help modulate immune responses in JIA and beyond.
Tumor-derived colorectal cancer organoids induce a unique Treg cell population by directing CD4+ T cell differentiation. Aristin Revilla S, Fredericks CL, Prekovic S, Mocholi E, Kranenburg O, Coffer PJ. (2025)
Here we develop a novel in vitro co-culture model to investiagate interactions between colorectal tumors and Treg cells. Our data demonstrate that tumor-organoids can directly induce CD4+ differentiation to generate Treg cells that are transcriptionally similar to those found in CRC patients.
Pyruvate metabolism controls chromatin remodeling during CD4+ T cell activation. Mocholi E, Russo L, Gopal K, Ramstead AG, Hochrein SM, Vos HR, Geeven G, Adegoke AO, Hoekstra A, van Es RM, Pittol JR, Vastert S, Rutter J, Radstake T, van Loosdregt J, Berkers C, Mokry M, Anderson CC, O'Connell RM, Vaeth M, Ussher J, Burgering BMT, Coffer PJ. (2023)
These data demonstrate the tight integration of metabolic and histone-modifying enzymes, allowing metabolic reprogramming to fuel CD4+ T cell activation. Targeting this pathway may provide a therapeutic approach to specifically regulate antigen-driven T cell activation.
Regulation of a progenitor gene program by SOX4 is essential for mammary tumor proliferation. Roukens MG, Frederiks CL, Seinstra D, Braccioli L, Khalil AA, Pals C, De Neck S, Bornes L, Beerling E, Mokry M, de Bruin A, Westendorp B, van Rheenen J, Coffer PJ. (2021)
Our study identifies a novel role for SOX4 in maintaining mammary tumors in an undifferentiated and proliferative state. Therapeutic manipulation of SOX4 function could provide a novel strategy for cancer differentiation therapy, which would promote differentiation and inhibit the cycling of tumor cells.
C/EBPα is a crucial determinant of epithelial maintenance by preventing epithelial-to-mesenchymal transition. Lourenço, A., Roukens, G.,Seinstra, D., Frderiks, C., Pals, C., Vervoort, S.J., Margarido, A.S., van Rheenen, J. and Coffer, P.J. (2020)
Our data suggest that C/EBPα is a master epithelial “gatekeeper” whose expression is required to prevent unwarranted epithelial-to-mesenchymal transition (EMT). Controlling C/EBPα expression may provide a way to prevent or reduce metastasis.
Nemo-like kinase drives Foxp3 stability and is critical for maintenance of immune tolerance by regulatory T cells. Fleskens, V., Minutti, C., Wu, X., Wei, P., Pals, C.E.G.M., McCrae, J., Hemmers, S., Groenewold, V., Rudensky, A., Pan, F., Li, H., Zaiss, D. and Coffer, P.J. (2019)
Here we demonstrate that TCR-mediated signaling can regulate the NLK protein kinase to sustain Foxp3 transcriptional activity through the stabilization of protein levels, thereby maintaining Treg cell suppressive function. This pathway may provide a novel therapeutic avenue for reversibly modulating Treg cell functionality.
Forkhead transcription factors as context-dependent regulators of lymphocyte homeostasis. Zaiss, D. and Coffer, P.J. (2018)
Nature Reviews Immunology 18, 703-715
Focusing on the various mechanisms by which their functional activity is modulated, as well as on their mechanisms of action, we discuss how specific Forkhead transcription factors control lymphocyte function, allowing for the establishment and maintenance of immune homeostasis.
SOX4-SMAD3 interaction redirects TGF-β-mediated transcriptional output in a context-dependent manner. Vervoort, S.J., Lourenco, A.R., Tufegdzic-Vidakovic, A., Sandoval, J.L., Rueda, O.M., Frederiks, C., Russell, R., Caldas, C., Bruna, A. and Coffer, P.J. (2018)
Nucleic Acid Research 46, 9578-9590
These findings identify SOX4 as an important SMAD3 co-factor controlling transcription of pro-metastatic genes and context-dependent shaping of the cellular response to TGF-β. Targeted disruption of the interaction between these factors may have the potential to disrupt pro-oncogenic TGF-β signaling, thereby impairing tumorigenesis.
Autophagy is a tolerance-avoidance mechanism that modulates TCR-mediated signaling and cell metabolism to prevent the induction of T cell anergy. Mocholi, E., Dowling, S.D., Botbol, Y., Gruber, R.C., Ray, A.K., Vastert, S., Shafit-Zagardo, B., Coffer, P.J. and Macian, F. (2018)
In response to activation, CD4+ T cells upregulate autophagy, however, the functional consequences of this have not been fully elucidated. Our data show that inhibition of autophagy during CD4+ T cell activation induces a long-lasting state of hypo-responsiveness that is accompanied by the expression of an anergic gene signature. We propose that autophagy constitutes a tolerance-avoidance mechanism, which determines CD4+ T cell fate.
Global transcriptional analysis identifies a novel role for SOX4 in tumor-induced angiogenesis. Vervoort, S.J., de Jong, O.G., Vermeulen, J.F., Bella, L., Lourenco, A-R., Frederiks, C., Tufegdzic-Vidakovic, A., Russell, R., Moelans,C., van Amersfoort, M., Dallman, M.J., Bruna, A., Caldas, C., Nieuwenhuis, E., van der Wall, E., Derksen, P., van Diest, P., Lam, W. W-F., Verhaar, M., Mokry, M. and Coffer, P.J.
The expression of the transcription factor SOX4 is increased in many human cancers, however, the pro-oncogenic capacity of SOX4 can vary greatly depending on the type of tumor. These findings provide novel mechanistic insights into context-dependent SOX4 target gene selection, and uncover a novel pro-oncogenic role for this transcription factor in promoting tumour-induced angiogenesis.
Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. (2013)
The delicate balance between pro- and anti-inflammatory mechanisms, essential for gut immune homeostasis, is affected by the composition of the commensal microbial community. Here, results suggest that bacterial metabolites mediate communication between the commensal microbiota and the immune system, affecting the balance between pro- and anti-inflammatory mechanisms.
Stabilization of the Transcription Factor Foxp3 by the Deubiquitinase USP7 Increases Treg-Cell-Suppressive Capacity. van Loosdregt, J., Fleskens, V., Fu, J., Brenkman, A.J., Bekker, C.P.J., Pals, C.E.G.M., Meerding, J., Berkers, C.R., Barbi, J., Grone, A., Sijts, A.J.M., Maurice, M.M., Kalkhoven, E., Prakken, B.J., Ovaa, O., Pan, F., Zaiss, D.M.W. and Coffer, P.J. (2013)
Here, we demonstrate that the expression of the Treg master transcription factor Foxp3 can be regulated through the polyubiquitination of multiple lysine residues, resulting in proteasome-mediated degradation. Our data reveal a molecular mechanism in which rapid temporal control of Foxp3 expression in Treg cells can be regulated by ubiquitination, thereby modulating Treg cell numbers and function. Regulating Foxp3 ubiquitination may provide a way of controlling immune homeostasis. See also News & Views: Laurence et al, 2013. Immunity 39, 201-203
Canonical Wnt signaling negatively modulates T regulatory cell function. van Loosdregt, J., Fleskens, V., Tiemessen, M.T., Mokry, M., van Boxtel, R., Meerding, J., Pals, C.E.G.M., Kurek, D., Baert, M.R., Delmarre, E.M., Grone, A., Groot-Koerkamp, M.J., Sijts, A.A.M., Maurice, M.M., van Es, J.H., ten Berge, D., Holstege, F.C., Staal, F.J.T., Zaiss, D.M.W., Prakken, B.J. and Coffer, P.J. (2013)
Here, we demonstrate that T cell factor 1 (TCF1) and Foxp3 associate in Treg cells, and that active Wnt signaling disrupts Foxp3 transcriptional activity. We propose a model in which Wnt produced under inflammatory conditions represses Treg cell function, allowing a productive immune response, but, if uncontrolled, could lead to the development of autoimmunity.
Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. van der Vos, K.E., Eliasson, P., Proikas-Cezanne, T., Vervoort, S.J., van Boxtel, R., Putker M., van Zutphen, I.J., Mauthe, M., Zellmer, S., Pals, C., Verhagen, L.P., Groot-Koerkamp, M.J., Braat, A.K., Dansen, T.B., Holstege, F.C., Gebhardt, R., Burgering, B.M., Coffer, P.J. (2012)
Nature Cell Biology 14, 829-37
Here we report an unexpected role for FOXO transcription factors in regulating autophagy by modulating intracellular glutamine levels. These findings reveal a growth-factor-responsive network that can directly modulate autophagy through the regulation of glutamine metabolism. See also News & Views: Sandri, 2012. Nature Cell Biology 4, 786-788